Research overview
The attachment of the small protein, ubiquitin, to target proteins regulates a vast array of biological processes. We are interested in the role of ubiquitination in regulating transcription and the DNA damage response. We study the protein complexes that attach, recognize and remove ubiquitin modifications, as well as the way in which cross-talk between ubiquitination and other post-translational modifications regulates chromatin activity. We use x-ray crystallography, cryo-EM, enzymology, cell-based assays and a variety of biophysical tools to gain insights into the mechanisms underlying these essential cellular processes. Below is a summary of ongoing research projects:
The role of histone H2B ubiquitination in transcription and chromatin dynamics
Monoubiquitination of histone H2B (H2B-Ub) plays a role in transcription, DNA 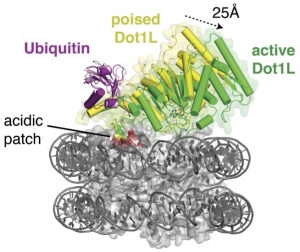 replication and the DNA damage response. H2B-Ub is a rapidly turned over histone mark that is found in actively transcribed regions. H2B-Ub plays a role in recruiting the histone chaperone, FACT, which mediates both disassembly and assembly of nucleosomes during transcription and DNA replication. Monoubiquitinated H2B also activates histone methyltransferases that methylate lysine residues in histone H3 at K4 and K79, which are marks of actively transcribed genes. We are interested in the mechanism by which the attachment of ubiquitin to histone H2B promotes transcription activation.
replication and the DNA damage response. H2B-Ub is a rapidly turned over histone mark that is found in actively transcribed regions. H2B-Ub plays a role in recruiting the histone chaperone, FACT, which mediates both disassembly and assembly of nucleosomes during transcription and DNA replication. Monoubiquitinated H2B also activates histone methyltransferases that methylate lysine residues in histone H3 at K4 and K79, which are marks of actively transcribed genes. We are interested in the mechanism by which the attachment of ubiquitin to histone H2B promotes transcription activation.
Rahman S, Hoffmann NA, Worden EJ, Smith ML, Namitz KEW, Knutson BA, Cosgrove MS, Wolberger C. Multistate structures of the MLL1-WRAD complex bound to H2B-ubiquitinated nucleosome. (2022) Proc Natl Acad Sci U S A. 119(38):e2205691119. doi: 10.1073/pnas.2205691119.
Worden EJ, Zhang X, Wolberger C. (2020) Structural basis for COMPASS recognition of an H2B-ubiquitinated nucleosome. Elife 9. pii: e53199. doi: 10.7554/eLife.53199.
Worden EJ, Hoffmann N, Hicks C, Wolberger C (2019) The mechanism of cross-talk between histone H2B ubiquitination and H3 methylation by Dot1L. Cell 176(6):1490-1501. doi: 10.1016/j.cell.2019.02.002
Nune, Morgan, Connell, McCullough, Jbara, Sun, Brik, Formosa, Wolberger (2019) FACT and Ubp10 collaborate to modulate H2B deubiquitination and nucleosome dynamics. eLife pii: e40988. doi: 10.7554/eLife.40988
SAGA: Ubiquitination and acetylation in gene activation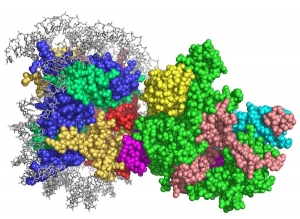
The 1.9 MDa yeast SAGA complex has been a paradigm for understanding the connection between histone modifications and gene activation as well as the cross-talk between different types of modifications. SAGA functions in a cascade of posttranslational modifications that begins with monoubiquitination of histone H2B-K123m which triggers methylation of histone H3-K4. The SAGA complex is recruited, where it deubiquitinates H2B, recognizes the H3K4 trimethyl mark and acetylates histone H3. The 19 SAGA subunits are conserved from yeast to humans and are organized into subcomplexes including the four-protein SAGA deubiquitinating module (DUBm) and the four-protein SAGA histone acetyltransferase (HAT) module. Our structural and biochemical studies are aimed at uncovering how SAGA recognizes its nucleosomal substrates and how the multiple histone modifying activities of SAGA are coordinated during the process of gene activation. We are also using the insights from our structural studies to identify highly specific inhibitors of the SAGA deubiquitinating module.
Morgan M, Ikenoue T, Suga H, Wolberger C. (2022) Potent macrocycle inhibitors of the human SAGA deubiquitinating module. Cell Chem Biol. 29(4):544-554.e4. doi: 10.1016/j.chembiol.2021.12.004.
Morgan, Haj-Yahya, Ringel, Bandi, Brik, Wolberger (2016) Structural basis for histone H2B deubiquitination by the SAGA DUB module. Science 351: 725-8. PubMed
Ringel, Cieniewicz, Taverna, Wolberger (2015) Nucleosome competition reveals processive acetylation by the SAGA HAT module. PNAS 6;112(40):E5461-70. PubMed
Samara, Ringel, Wolberger (2012) A role for intersubunit interactions in maintaining SAGA deubiquitinating module structure and activity. Structure 20:1414-24. PubMed
Samara, Datta, Berndsen, Zhang, Yao, Cohen, Wolberger C. (2010) Structural insights into the assembly and function of the SAGA deubiquitinating module. Science 328: 1025-9. PubMed
Ubiquitin signaling in the DNA damage response
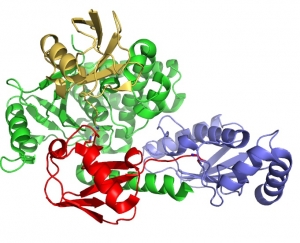
Attachment of ubiquitin or the ubiquitin-like protein, SUMO, plays a critical role in the response to DNA damage. Monoubiquitin, K63-linked polyubiquitin, K48-linked polyubiquitin and mixed SUMO-ubiquitin chains play distinct roles in signaling DNA damage and recruiting the necessary enzymes needed for DNA repair. We study the enzymes that specifically assemble and disassemble ubiquitin signals at damage sites, as well as the proteins that recognize different types of chains as distinct signals.
Lombardi PM, Haile S, Rusanov T, Rodell R, Anoh R, Baer JG, Burke KA, Gray LN, Hacker AR, Kebreau KR, Ngandu CK, Orland HA, Osei-Asante E, Schmelyun DP, Shorb DE, Syed SH, Veilleux JM, Majumdar A, Mosammaparast N, Wolberger C. (2022) The ASCC2 CUE domain in the ALKBH3-ASCC DNA repair complex recognizes adjacent ubiquitins in K63-linked polyubiquitin. J Biol Chem. 298(2):101545. doi: 10.1016/j.jbc.2021.101545.
Brickner, Soll , Lombardi , Vågbø , Mudge, Oyeniran, Rabe , Jackson, Sullender, Blazosky, Byrum, Zhao, Corbett, Gécz, Field, Vindigni, Slupphaug, Wolberger, Mosammaparast (2017) A ubiquitin-dependent signalling axis specific for ALKBH-mediated DNA dealkylation repair. Nature 551: 389-393.
Wiener, DiBello, Lombardi, Guzzo, Zhang, Matunis, Wolberger (2013) E2 ubiquitin-conjugating enzymes regulate the deubiquitinating activity of OTUB1. NSMB 20:1033-9. PubMed
Guzzo, Berndsen, Zhu, Gupta, Datta, Greenberg, Wolberger, Matunis (2012) RNF4-dependent hybrid SUMO-ubiquitin chains are signals for RAP80 and thereby mediate the recruitment of BRCA1 to sites of DNA damage. Sci Signal. 5(253):ra88. PubMed
Wiener, Zhang, Wang, Wolberger (2012) The mechanism of OTUB1-mediated inhibition of ubiquitination. Nature 483:618-22. PubMed
